How Proteins Fold Into Their Critical Shapes
Experimental evidence provided by a Cornell researcher and colleagues at the Scripps Research Institute in La Jolla, Calif., support a long-held theory of how and where proteins fold to create their characteristic shapes and biological functions.
The theory proposes that proteins start to fold in specific places along an amino acid chain (called a polypeptide chain) that contains nonpolar groups, or groups of molecules without a charge, and continue to fold by aggregation, i.e., as several individuals of these nonpolar groupings combine. Using the same principle that separates oil and water, these molecules are hydrophobic - they avoid water and associate with each other.
In the water-based cell fluid, where long polypeptide chains are manufactured and released by ribosomes, the polypeptide chains rapidly fold up into their biologically functional structure. The theory proposes that there are sites along the polypeptide chains where hydrophobic groups initially fold in on themselves, creating small nonpolar (hydrophobic) pockets that are protected from the water.
"What drives this polypeptide chain to fold up?" asked Harold Scheraga, professor emeritus of chemistry and chemical biology at Cornell and a co-author of a paper published in the Aug. 29 issue of the Proceedings of the National Academy of Sciences (and available online). "That has been the subject of my investigations for some time, and the cited experimental verification of the theory provides a sound basis for further computational work to identify the specific steps in the folding pathway.
"Protein folding is a frontier problem in protein chemistry," said Scheraga, noting that an ability to predict how and where proteins fold could lead to understanding such protein misfolding diseases as Alzheimer's and cystic fibrosis, designing drugs that act on proteins and even creating designer proteins with new functions.
The theory is based on two methods to show that initial folding sites occur among nonpolar groups in a polypeptide chain. Lead author H. Jane Dyson and Peter Wright, both professors of molecular biology at the Scripps Research Institute, used an experimental nuclear magnetic resonance procedure to validate the predicted results of the two theoretical methods.
The first method used supercomputers to calculate the energy required to convert a polypeptide chain into a collapsed hydrophobic pocket. The folds occur in several places that require the least possible energy to maintain. By finding these places where the nonpolar groups exist, the researchers better understand where folding occurs along a linear polypeptide chain.
The second method involved mapping a folded protein by tracing the folding steps required to arrive at the protein's native structure. This method mapped three stages of folding. First, the short-range contacts between amino acids that are very close to each other were mapped, revealing the initial nonpolar (hydrophobic) folds. The next two stages show folds that occur between points that are farther from each other along the polypeptide chain. These secondary folds may attach two or three hydrophobic pockets.
These two methods were used together in this study to pinpoint where on a polypeptide chain the nonpolar segments occur and where initial folding takes place and then propagates to the final folded form.
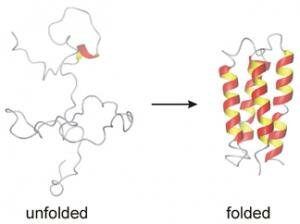
"This illustration shows a designed protein transformed from an unfolded into a folded form. (Credit: Adam Liwo / Courtesy of Cornell University)"
Source: Cornell University
|